| Step |
Description |
View |
| 1 |
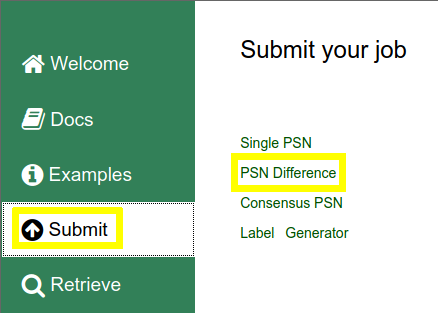
Click on Submit in the vertical menu on the left and then on PSN Difference link on the right |
 |
| 2 |
-
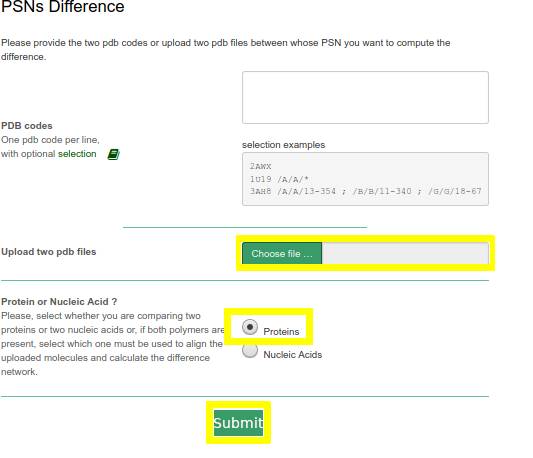
In this case example, structural differences related to arrestin activation will be inferred by computing the PSN difference between the crystal structures of the inactive (PDB: 1CF1) and active (PDB: 5W0P) states of arrestin-1.
- Upload two pdb files (download the files used in this example: 1CF1 and 5W0P)
- Select Proteins option
- Click on Submit
|
 |
| 3 |
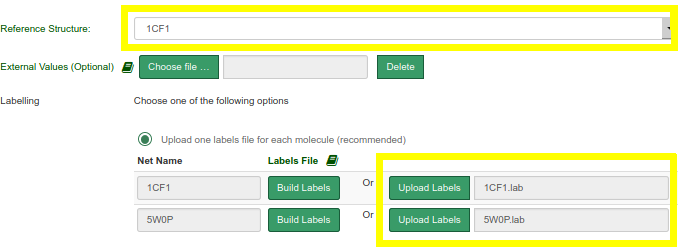
- Select 1CF1 as reference structure
- Upload one labels file for each molecule (download the files used in this example: 1CF1 and 5W0P)
- Click on Submit at the bottom of the page
|
 |





