| Step |
Description |
View |
| 1 |
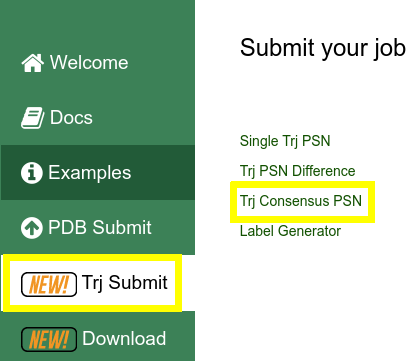
Click on Trj Submit in the vertical menu on the left and then on PSN Difference link on the right |
 |
| 2 |
-
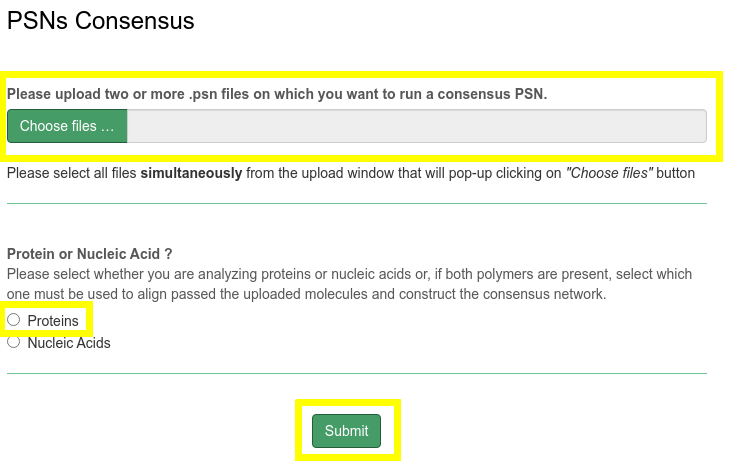
In this case example, network features shared in common by the inactive (PDB 3LNX) and active (PDB 3LNY) states of second PDZ domain from human PTP1E activation will be inferred calculating a consensus PSN.
- Download the two .psn files 3LNX and 3LNY used in this example and upload them
- Select Proteins option
- Click on Submit
|
 |
| 3 |
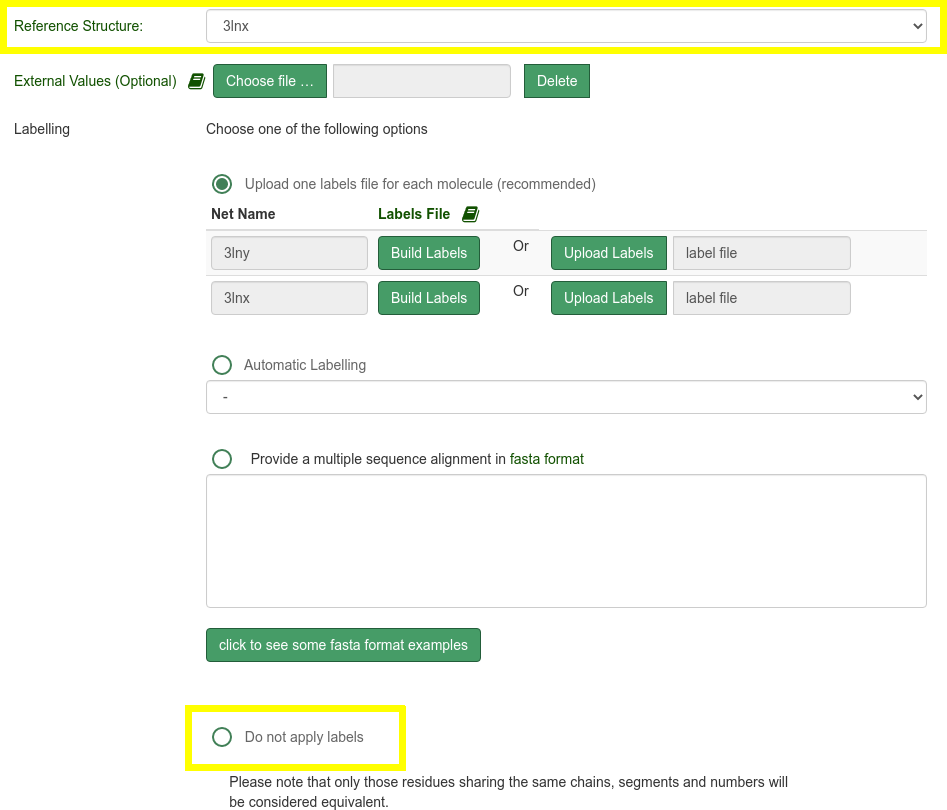
- Select 3lnx as reference structure
- Select "Do not apply labels" option
- Click on Submit at the bottom of the page
|
 |





